Full Length Research Article - (2022) Volume 17, Issue 5
*Correspondence: Mustafa Salah Hasan, University of Fallujah, College of Veterinary Medicine, Iraq, Email:
2University of Babylon/DNA Research Center, Iraq
3College of Health and Medical Technology, Al-Zahraa University for Women, Karbala, Iraq
4University of Fallujah, College of Veterinary Medicine, Iraq
Received: 05-Sep-2022 Accepted: 26-Sep-2022 Published: 26-Sep-2022
Abstract
The aim of this study was molecular detection of opportunistic bacteria Pantoea spp. From different human samples. From June 2021 to June 2022, 141 clinical samples were gathered from clinics throughout the Babylon province. Swabs were taken from various clinical samples, including 28 samples from the pharynx, ears (26), stool (22), wounds (18), pus (15), cough (17) and burns (16). These samples were cultured on primary medium, then biochemical tests were done for identification of bacteria. The DNA of bacteria were extracted by using kit purchased from (Promega, Madison). Pantoea species were identified using gene 16rsRNA primers. By bacteriological methods, out of 140 samples, 6 isolates were suspected as Pantoea spp. On MacConkey agar, smooth, convex, punctate, umbilicated fermenting lactose, shining colonies were produced, after Gram staining revealed Gram negative rods. The molecular way demonstrates that the isolates are from Pantoea species. During this investigation, the PCR method has a 100% sensitivity rate for isolating isolates.
In conclusion, Pantoea spp. was isolated from different human sources in significant percentage, which may be causative agent for different diseases.
Keywords
Sport Psychology, Exercise, Molecular, Pantoea spp., Human
Introduction
Various hosts, including plants, animals, insects, as well as humans, are often found in close proximity to Pantoea strains [1,2]. Although several Pantoea species are well-known plant pathogens [3-5], they have also been identified from clinical samples. Pantoea agglomerans has been identified from the blood of children suffering from bacteremia, septicemia, peritonitis, osteomyelitis, septic arthritis, pneumonia, as well as septic arthropathy [6]. Cases of P. agglomerans in humans are often the consequence of hospital-acquired infections or contamination of a wound by plant matter [7]. Isolates of other species, including as Pantoea eucalypti,Pantoea ananatis, Pantoea dispersa, as well as Pantoea septica, have also been found in many clinical sources,such as blood, wounds, CSF, faeces, skin, cysts, abscesses, fractures, as well as urethra & trachea [2].
Additionally, pantoea has been linked to several epidemics that have killed newborns [8,9]. The pathogenic potential of some Pantoea species for humans is still being challenged [10], despite data showing that many clinical strains are not Pantoea at all. Many strains that were formerly thought to be members of the Pantoea genus have been reassigned to other genera as a consequence of taxonomic and nomenclatural changes [11]. Moreover, it is difficult to place Pantoea strains into a species group using solely their metabolic profile, which has led to the misidentification of some Pantoea isolates[12].
The enterobacteriaceae family, which does not stain positively on the Gram stain, is known to include the bacteria Pantoea spp. [13]. The primary issue is that, although not being a mandatory infection, Pantoea spp. bacteria are linked to opportunistic illnesses [14].
The cellulase enzyme, for instance, is useful for studying the cellulose polymer that makes up the walls of plant cells, which both defends the plant against invasion and makes the bacterium more dangerous. The genes responsible for the synthesis of exopolysaccharide (EPS) in Pantoea spp. [15] have been identified, and EPS plays an important role in the adherence and pathogenicity of Pantoea spp. The regulatory genes rcsA and rcsB regulate the synthesis of the exopolysaccharide known as capsule polysaccharide (EPS) [16].
Producing more pilius is facilitated by the hpaA gene, which in turn improves host pathogenicity [17] by making bacteria more adept at sticking to host tissue and interfering with cellular host function.
The aim of this study was molecular detection of opportunistic bacteria Pantoea spp. From different human samples.
Materials and Methods
From June 2021 to June 2022, 141 clinical samples were gathered from clinics throughout the Babylon province. Swabs were taken from various clinical samples, including 28 samples from the pharynx, ears (26), stool (22), wounds (18), pus (15), cough (17) and burns (16). These samples were cultured on primary medium, then biochemical tests were done for identification of bacteria [18].
The DNA of bacteria were extracted by using kit purchased from (Promega, Madison). Pantoea species were identified using gene 16rsRNA primers. The gene sequence was:
F CCTGGACAAAGACTGACGCT, R CGCTTCTCTTTGTATGCGCC The reaction was carried out in 34 cycles, each lasting 1 minute at 95°C, 45 seconds at 53°C, and 2 minutes at 72°C after a 5-minute initial denaturation. Five l of the PCR product were run on 1% (w/v) agarose gel after PCR amplification, stained with ethidium bromide, and examined under a UV transilluminator.
Results and Discussions
By bacteriological methods, out of 140 samples, 6 isolates were suspected as Pantoea spp. On MacConkey agar, smooth, convex, punctate, umbilicated fermenting lactose, shining colonies were produced, after Gram staining revealed Gram negative rods.
By using API 20E assays, the Pantoea spp. were biochemically described. All six isolates fit the description of the Pantoea genus. They lacked urease, did not decarboxylate lysine or ornithine, and did not create H2S from thiosulfat. To the best of the API 20E system's knowledge, no species-level identifications have been made with any of the isolates. However, only Pantoea species are included in the current database; these findings have been verified in the past by [19].
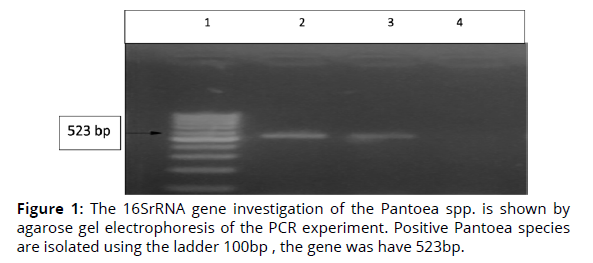
In this work, the isolates of Pantoea spp. were identified by using of gene 16SrRNA. The findings in Fig. 1 demonstrate that the gene 16SrRNA, which is the diagnostic gene responsible for this bacteria, is present in all isolates and has a molecular weight of (523 bp). This demonstrates that the isolates are from Pantoea species. During this investigation, the PCR method has a 100% sensitivity rate for isolating isolates (Figure 1).
The 16S rRNA gene was used to screen 18 isolates for Pantoea spp., and 9 of the isolates were found to be members of this bacterial family, as reported in the study [20]. A study of 21 isolates of the genus Pantoea found that 95.6% had the 16S rRNA gene, making them potentially dangerous to humans and other animals [21].
Conclusion
Pantoea spp. was isolated from different human sources in significant percentage, which may be causative agent for different diseases.
References
Nadarasah G, Stavrinides J. Quantitative evaluation of the host-colonizing capabilities of the enteric bacterium Pantoea using plant and insect hosts. Microbiol (United Kingdom). 2014;160: 602–615.
Walterson AM, Stavrinides J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev. 2015;39: 968–984.
Roper MC. Pantoea stewartii subsp. stewartii: Lessons learned from a xylem-dwelling pathogen of sweet corn. Mol Plant Pathol. 2011;12: 628–637.
Brady CL, Venter SN, Cleenwerck I, Engelbeen K, Vancanneyt M, Swings J, et al. Pantoea vagans sp. nov., Pantoea eucalypti sp. nov., Pantoea deleyi sp. nov. and Pantoea anthophila sp. nov. Int J Syst Evol Microbiol. 2009;59: 2339–2345.
Coutinho TA, Venter SN. Pantoea ananatis: An unconventional plant pathogen. Mol Plant Pathol. 2009;10: 325–335.
Cruz AT, Cazacu AC, Allen CH. Pantoea agglomerans, a plant pathogen causing human disease. J Clin Microbiol. 2007;45: 1989–1992.
Dutkiewicz J, Mackiewicz B, Lemieszek MK, Golec M, Milanowski J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Ann Agric Environ Med. Institute of Rural Health; 2016;23: 197–205.
Van Rostenberghe H, Noraida R, Wan Pauzi WI, Habsah H, Zeehaida M, Rosliza AR, et al. The clinical picture of neonatal infection with Pantoea species. Jpn J Infect Dis. 2006;59: 120–121.
Bergman KA, Arends JP, Schölvinck EH. Pantoea agglomerans septicemia in three newborn infants. Pediatr Infect Dis J. 2007;26: 453–454.
Rezzonico F, Smits THM, Duffy B. Misidentification slanders Pantoea agglomerans as a serial killer. J Hosp Infect. 2012;81: 137–139.
Rezzonico F, Smits TH, Montesinos E, Frey JE, Duffy B. Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 2009;9 10.1186/1471-2180-9-204.
Rezzonico F, Stockwell VO, Tonolla M, Duffy B, Smits THM. Pantoea clinical isolates cannot be accurately assigned to species based on metabolic profiling. Transpl Infect Dis. 2012;14: 220–221.
Leonila M.L. Acioly1, Vilar J. Carlos3, Aline Barbosa da Silveira2, Fabiola C. Gomes de Almeida4, Thayse Alves de Lima e Silva4 and Galba Maria de Campos-Takaki.(2017). Isolation, Identification, Characterization and Enzymatic Profile of the New Strain of Pantoea agglomerans. International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 6 Number 11 (2017) pp. 4152-4163.
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., & Milanowski, J. (2016). Pantoea agglomerans: a mysterious bacterium of evil and good. Part III. Deleterious effects: infections of humans, animals and plants. Annals of Agricultural and Environmental Medicine, 23(2).
Schwarz, S., Hood, R. D., & Mougous, J. D. (2010). What is type VI secretion doing in all those bugs?. Trends in microbiology, 18(12), 531-537.
Majdalani, N., and Gottesman, S. (2005). The Rcs phosphorelay: A complex signal transduction system. Annu. Rev. Microbiol. 59, 379-40The molecular basis for transformation of an epiphyte into a gall-forming pathogen as exemplified by Erwinia herbicola pv. gypsophilae. Pages 19-52 in: Plant–Microbe Interactions. Vol. 6. G. Stacey and N. Keen, eds. American Phytopathological Society, St. Paul, MN, U.S.A.
Lu H, Patil P, Van Sluys MA, White FF, Ryan RP, Dow JM, Rabinowicz P, Salzberg SL, Leach JE, Sonti R, et al. (2008). Acquisition and evolution of plant pathogenesis–associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS ONE. 3:e3828.
Markey, B., Leonard, F., Archambault, M., Cullinane, A., & Maguire, D. (2013). Clinical veterinary microbiology e-book. Elsevier Health Sciences.
Holt JG,Krieg NR,Snaith PA,Staley JT,and Williams ST.Bergey’s manual of Determinative Bacteriology. Lippincott Williams and wilkins,Philadelphia. (2000);ISBN-683.
Cheng, A., Liu, C. Y., Tsai, H. Y., Hsu, M. S., Yang, C. J., Huang, Y. T., ... & Hsueh, P. R. (2013). Bacteremia caused by Pantoea agglomerans at a medical center in Taiwan, 2000– 2010. Journal of Microbiology, Immunology and Infection, 46(3), 187-194.
Deletoile, A., Decre, D., Courant, S., Passet, V., Audo, J., Grimont, P., & Brisse, S. (2009). Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. Journal of Clinical Microbiology, 47(2), 300-310.