Research - (2022) Volume 17, Issue 3
Molecular Detection of Pophyromonas gingivalis and Some Interleukins Related with Periodontal Inflammatory Diseases
Natheer Ahmad Hameed1*, Reyam Abdul Khuder Mohammed2, Thaar Mohammed Najim3 and Mustafa Salah Hasan4*Correspondence: Natheer Ahmad Hameed, Almaarif Univerity College, Department of Nursing, Iraq,
Abstract
In this study, a total of 50 abscess swabs were collected from patients suffering from periodontal inflammatory diseases, the result showed that, out of 50 specimens, 33(66%) were positive culture on different types of media and 17(34%) were related to other types of causative agents (no growth on culture media). According to positive cultures, gram stain, biochemical tests and Vitek system, 9(27.2%) Pophyromonas gingivalis out of 33 specimens was considered main types of bacterial isolates were related with periodontal inflammatory diseases, while 24(72.2%) was related to other types of bacteria. Molecular detection of specific 16srRNA gene of Pophyromonas gingivalis were done in all Pophyromonas gingivalis isolates from periodontal inflammatory diseases samples, the results showed that, all of 9(100%) of Pophyromonas gingivalis isolates were gave positive results for this gene by the presence of (153bp) bands when compared with allelic ladder. In addition, molecular study of specific 16srRNA gene of IL-6 was done for all 9 isolates that previously detected as Bifidobacterium. Polymerase chain reaction is a sensitive and specific method for identification of this gene. The results showed that, all 9(100%) Pophyromonas gingivalis isolates were gave positive results for this gene by the presence of (174bp) bands when compared with allelic. However, molecular detection of specific 16srRNA gene of IL-10 were done in all Pophyromonas gingivalis isolates from periodontal inflammatory diseases, samples, the results showed that, all of 9(100%) of Pophyromonas gingivalis isolates were gave positive results for this gene by the presence of (627bp) bands when compared with allelic ladder. The mean differences IL-6 and IL-10 levels between study groups including (patients with periodontal inflammatory diseases and control group) were significant increase (P≤ 0.05) in IL-6 and IL-10 in patients with periodontal inflammatory diseases compared with control group.Aim to study
The aimed to detection of Pophyromonas gingivalis and interleukins genes (IL-6, IL-10) that related to periodontal inflammatory diseases.
Keywords
Periodontal inflammatory diseases. Interleukins. Genes. PCR
Introduction
Inflammation is a special state in terms of tissue response to injury that causes a shift in its metabolic status from an anabolic towards a catabolic one [1]. In the strictest sense, these characteristics differ from that of a tissue degenerative process firstly, by the rate with which catabolism exceeds anabolism and, secondly, the rise in the osmotic pressure causing significant fluid accumulation. Therefore, inflammation is almost a pathological process arising as a direct consequence of a tissue insult or injury. To counterbalance it, an opposing reactive process, the anti-inflammatory response, is characterized by proteolysis inhibition [2]. In a healthy state, after inflammation reaches its maximum capacity, an antiinflammatory response ensues to restore tissue homeostasis, representing a true defensive response [3]. Thus, inflammation ensued by an antiinflammatory response is vital for complete healing which, if not the case, can develop into chronic inflammation and autoimmune diseases [4]. Based on onset and duration, inflammation can be categorized into acute and chronic [5]. Acute inflammation is characterized by a rapid onset, short duration, and profound signs and symptoms. Acute inflammation occurs through several stages that are recognized through cellular changes [6]. Initially, there is an increase in blood flow within and around the vasculature at the site of injury. An increase in vascular permeability allows recruited immune cells extravasate into the injured parenchyma where they migrate to the injured cells using gradients of inflammatory molecules via chemotaxis [7]. Upon reaching the site of injury, immune cells proceed to phagocytose microbes and cellular debris which may be present [8]. The ultimate goal of the acute inflammatory response is clearance of damaged host cells and removal of pathogens. However, some recruited immune cells such as macrophages and dendritic cells travel to local lymph nodes where, as APCs, they process and present degraded phagocytosed debris advancing the immune response from that of an innate to an adaptive one [9]. In contrast, chronic inflammation is slow in onset and longer in duration with variable intensity of signs and symptoms. Chronic inflammation can follow acute inflammation, but it can also occur as a symptomless lowgrade, prolonged response to an inciting agent [10]. Most inflammatory diseases of the oral cavity involve the tissues of periodontium. The teeth emerge into the oral cavity after crossing bone and connective tissue as well as epithelial tissues. In the environment of the oral cavity, these hard structures are surrounded by a biofilm comprising of a vast collection of bacteria known to exist outside the colon. Such an architectural arrangement of soft and hard tissue compartments may or may not act jointly during the inflammatory responses [11]. The two common diseases affecting the health of the periodontium are gingivitis and PD. Gingivitis, or inflammation of the gingiva, is limited to the soft tissues comprising of epithelial and connective tissue [12]. The inflammatory changes in gingivitis are reversible with adequate hygiene measures. In the case of periodontal disease, the inflammatory process involves the periodontal ligament and alveolar bone. The progression from a reversible gingivitis to an irreversible loss of bone and ligamental tissues around the tooth has been described in terms of histopathological stages [13]. Periodontal health is likely to be challenged by a dysbiotic oral microbiota but ultimately, periodontal destruction is the result of a dysregulated immune response [14]. In healthy periodontal tissues, local immunity is orchestrated by the presence of antigen presenting cells (macrophages and dendritic cells), T lymphocytes (mostly CD4-positive helper T lymphocytes), and a comparatively lower number of neutrophils, B-cells and plasma cells. During inflammation, an increase is seen in most of these cells [15]. Inflammation is controlled by a host of extracellular mediators and regulators which include cytokines, chemokines and growth factors [16]. Cytokines are cellular messengers that are highly active within the environment of their cellular source (s). Structurally, they are synthesized as either proteins, glycoproteins or polypeptides. Majority of them have immunologically modifying effects. They are mostly secreted but may also be expressed on the cell membrane [17]. Chemokines are an excellent example of fast action inducing signal transduction. They are cytokines which are involved in chemotaxis, a process of attracting cells to a specific site via establishment of a chemically stimulating gradient. Cytokines play an important part in the host response to PD as they have putative roles in terms of tissue destruction or protection [18].
Materials and methods
Patients and collection of samples
The Cross Sectional study was carried out for a period of (1) year from March (2020) to March (2021). 100 patients were included in this study (50 patients were visit to dental clinic in Hilla city suffering from periodontal inflammatory diseases and 50 patients as healthy control). The sample were collected from abscess of each case by disposable cotton swabs at site of inflammation, and following standard procedure for microscopic examination and isolation of bacteria. Specimens were collected carefully to avoid any contamination. One aliquot of collected specimen was immediately inoculated in Blood agar media at bedside for aerobic culture. The rest of the specimen was transferred to the Department of Microbiology for further investigations, it was inoculated into Blood agar and Nutrient agar medium, then was incubated at (37oC) for (24) hours aerobically. Bacterial isolated were diagnosed by gram stain, colony morphology, biochemical test, Vitek 2 system and 16sr RNA specific gene for identification of Bifidobacterium. 3ml of blood samples were collected from each patient, then, separation of serum, the separation of the serum used for immunological study.
Ethical Approval
A valid consent was achieved from each patient before their inclusion in the study.
Questionnaire
A questionnaire was used to collect information from participating subjects regarding presence of self-reported chronic diseases and previous hospitalization history. Detailed information regarding oral health status, past dental history, oral hygiene measures, use of medication and smoking habits were recorded. Medical records and prescription for medication was requested for verifying self-reported conditions.
Inclusion criteria
18 years and above in age.
Exclusion criteria
Individuals were treatment with antibiotics in the last three months and / or undergoing treatment for periodontal disease during the last six months were excluded.
Identification of bacterial isolates by gram stain, biochemical tests
The identification tests, including cultural, morphological and biochemical characteristics were done for each isolate according to [19, 20].
Identification of bacterial isolates with Compact VITEK-2 System
All bacterial isolates were screened and identified via the Compact VITEK-2 System (BioMerieux). This is a phenotypic type of identification, which depends on biochemical reactions to identify the isolates. The Vitek-2 card contains 64 wells, which hold different fluorescent biochemical assays. Out of the 64, 20 carbohydrate assimilation were phosphatase, urea, nitrate, and actidione tests. The Vitek-2 machine controlled the card automatically including the filling, sealing, and then transferring the cards into the linked incubator (35°C). Each output report is decoded according to a particular algorithmic system. The acquired results were identified according to ID-GP (identification of Gram-positive bacteria) and ID-GN (identification of Gram-negative bacteria) databank. The ID results from these systems are proposed automatically by the respective accompanying software. The tests were repeated only if the initial results indicated “low discrimination” or “no ID”, and the repeat result was used for data analysis. All strains were inoculated onto culture media and then incubated overnight at 37°C. A single isolated colony was used for identification by the phenotypic VITEK-2 Systems method, done according to the manufacturer’s instructions (BioMerieux).
DNA extraction
This method was made according to the genomic DNA purification Kit supplemented by the manufacturing company Geneaid, (UK).
Detection of specific Pophyromonas gingivalis and interleukins genes
16sr RNA Specific genes primers sequences of Pophyromonas gingivalis and interleukins with amplicon size base pair (bp) and their condition were listed in table 1 (Table 1).
| Genes | Primer sequence (5ÃÂ?¹-3ÃÂ?¹) | Size of product bp | PCR condition | Reference |
|---|---|---|---|---|
| 16sr RNA of Pophyromonas gingivalis | F: 5′-GAG GAC CTT TGC CCA CCA-3′ R: 5′-GCG AAA ACT GAC CCT CG-3 |
153 | Step 1: 95ºC, 2 min. Step 2: 95ºC, 30 sec. Step 3: 57.0ºC, 30 sec. Step 4: 72ºC, 20.0 sec. Step 5: Repeat steps 2-4 29 more times Step 6: 72ºC, 5 min. Step 7: 4ºC, forever |
[20] |
| IL -6 | F: T G ACTT C AGCTTT ACT CTT GT R: CTGATTGGAACCCTTATTAAG |
174 | Step 1: 95ºC, 2 min. Step 2: 95ºC, 30 sec. Step 3: 57.0ºC, 30 sec. Step 4: 72ºC, 20.0 sec. Step 5: Repeat steps 2-4 29 more times Step 6: 72ºC, 5 min. Step 7: 4ºC, forever |
[22] |
| IL -10 | F: CTTAGGTCACAGTGACGTGG R: GTG AGC ACT ACCT G ACT AGC |
627 | Step 1: 95ºC, 2 min. Step 2: 95ºC, 30 sec. Step 3: 57.0ºC, 30 sec. Step 4: 72ºC, 20.0 sec. Step 5: Repeat steps 2-4 29 more times Step 6: 72ºC, 5 min. Step 7: 4ºC, forever |
[22] |
Statistical analysis
Statistical analysis was carried out using SPSS version 23. Continuous variables were presented as (Means ± SD). Student t-test was used to compare means between two groups.
Results
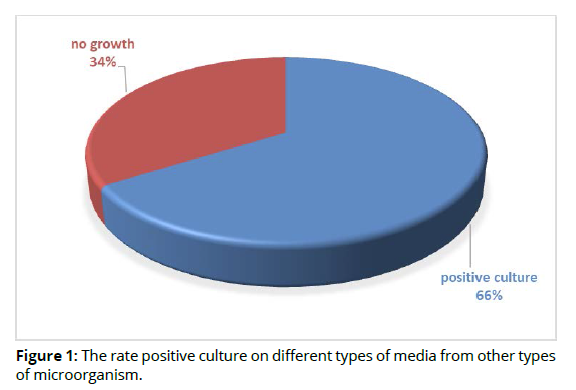
In this study, a total of 50 abscess swabs were collected from patients suffering from periodontal inflammatory diseases, the result showed that, out of 50 specimens, 33(66%) were positive culture on different types of media and 17(34%) were related to other types of causative agents (no growth on culture media) as shown in figure 1 (Figure 1).
According to positive cultures, gram stain, biochemical tests and Vitek system, 9(27.2%) Pophyromonas gingivalis out of 33 specimens was considered main types of bacterial isolates were related with periodontal inflammatory diseases, while 24(72.2%) was related to other types of bacteria as shown in table 2 (Table 2).
| Total No. of samples | Positive culture | Other types of causative agents | identification ofPophyromonas gingivalis | Other types of bacteria |
|---|---|---|---|---|
| 50 | 33 (66%) | 17(34%) | 9(27.2%) | 24(72.2%) |
| Total | 50 | Total: 33 | ||
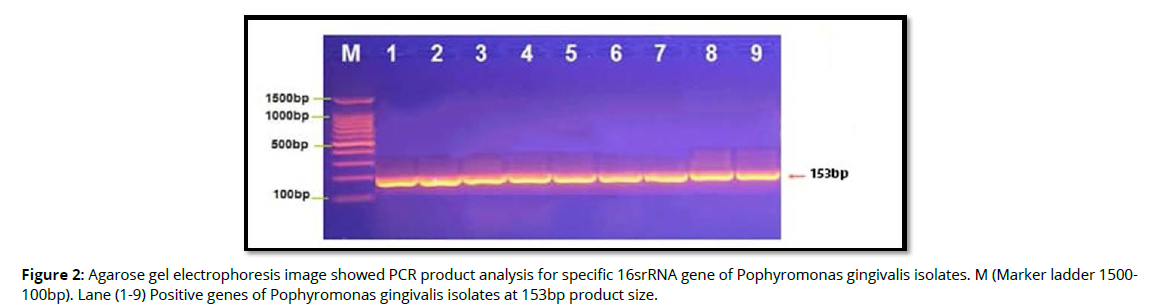
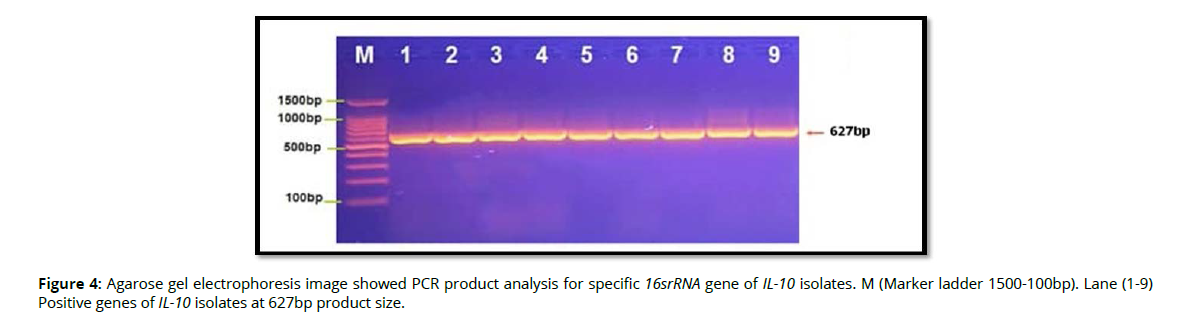
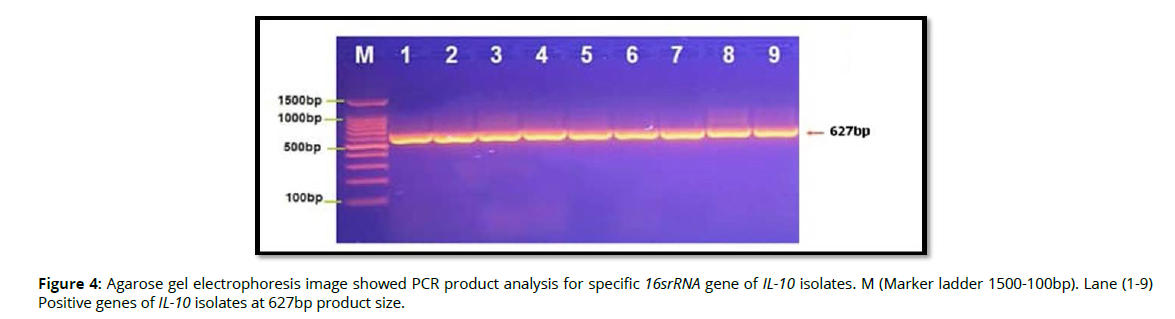
Molecular detection of specific 16srRNA gene of Pophyromonas gingivalis were done in all Pophyromonas gingivalis isolates from periodontal inflammatory diseases samples, the results showed that, all of 9(100%) of Pophyromonas gingivalis isolates were gave positive results for this gene by the presence of (153bp) bands when compared with allelic ladder as shown in figure 2 (Figure2). In addition, molecular study of specific 16srRNA gene of IL-6 was done forall 9 isolates that previously detected as Pophyromonas gingivalis. Polymerase chain reaction is a sensitive and specific method for identification of this gene. The results showed that, all 9(100%) Pophyromonas gingivalis isolates were gave positive results for this gene by the presence of (174) bp bands when compared with allelic ladder as shown in figure 3 (Figure 3). However, Molecular detection of specific 16srRNA gene of IL-10 were done in all Pophyromonas gingivalis isolates from periodontal inflammatory diseases, samples, the results showed that, all of 9(100%) of Pophyromonas gingivalis isolates were gave positive results for this gene by the presence of (627bp) bands when compared with allelic ladder as shown in figure 4 (Figure 4).
In this study, the mean differences IL-6 and IL-10 levels between study groups including (patients with periodontal inflammatory diseases and control group) was shown in table 3 (Table 3). The results showed that, there were significant increase (P≤ 0.05) in IL-6 and IL-10 in patients with periodontal inflammatory diseases compared with control group.
| Cytokine | Study groups | P-value | |
|---|---|---|---|
| patients with periodontal inflammatory diseases (N=50) | Healthy control (N=50) | ||
| IL-6 | 24.19 ± 1.42 | 9.45 ± 2.60 | P≤ 0.05 |
| IL-10 | 8.96 ± 3.58 | 4.11 ± 1.18 | P≤ 0.05 |
Discussion
Periodontal pathogens may include Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Peptostreptococcus micros, Pophyromonas gingivalis, Prevotella intermedia, Treponema denticola, Treponema forsythia, and putative periodontal pathogens such as Filifactor alocis and Parvimonas micra [23], especially organisms of the red complex (P. gingivalis, T. forsythia and T.
denticola)—though at least 17 new additional candidate organisms, including species or phylotypes from the phyla Bacteroidetes, Candidatus Saccharibacteria, Firmicutes, Proteobacteria, Spirochaetes, and Synergistetes, as well as possibly from the Archaea domain are reportedly associated with periodontal disease [24]. Porphyromonas gingivalis is a Gram-negative oral anaerobe that is involved in the pathogenesis of periodontitis and is a member of more than 500 bacterial species that live in the oral cavity. This anaerobic bacterium is a natural member of the oral microbiome, yet it can become highly destructive (termed pathobiont) and proliferate to high cell numbers in periodontal lesions: this is attributed to its arsenal of specialized virulence factors [25]. Porphyromonas gingivalis can locally invade periodontal tissues and evade the host defense mechanisms. In doing so, it utilizes a panel of virulence factors that cause deregulation of the innate immune and inflammatory responses [26]. The harsh inflammatory condition of the periodontal pocket suggests that this organism has properties that will facilitate its ability to respond and adapt to oxidative stress. Because the stress response in the pathogen is a major determinant of its virulence, a comprehensive understanding of its oxidative stress resistance strategy is vital [27]. Porphyromonas gingivalis is strongly correlated with chronic periodontitis. Its chronic persistence in the periodontium depends on its ability to evade host immunity without inhibiting the overall inflammatory response, which is actually beneficial for this and other periodontal bacteria. Indeed, the inflammatory exudate (gingival crevicular fluid) is a source of essential nutrients, such as peptides and heminderived iron [26]. Porphyromonas gingivalis contributes to the pathogenesis of aggressive periodontitis by inducing high levels of proinflammatory cytokines, such as IL-10 and IL-6 by peripheral CD4+ T helper cells [28]. Porphyromonas gingivalis, an etiological agent in severe forms of periodontitis, is a prominent component of the oral microbiome and a successful colonizer of the oral epithelium. This Gram-negative anaerobe can also exist within the host epithelium without the existence of overt disease [29]. Porphyromonas gingivalis seems to be a highly adapted pathogen of the oral microbiome. In recent years, investigations on the susceptibility factors of periodontitis have mainly focused on genes that modulate immunoregulation, such as cytokines, cell-surface receptors, chemokines, enzymes and proteins related to antigen recognition [30]. Cytokines, such as IL1A, IL1B, IL10 and IL6, are key factors that mediate the inflammatory process during periodontal disease. They have a role in B-cell activation, proliferation and differentiation, and are the majority of infiltrating cells in advanced periodontitis lesions because they may be responsible for the repeated cycles of tissue inflammation observed in these disorders [31].
Conclusion
Pophyromonas gingivalisis the one of the causative agent of related with periodontal inflammatory diseases, Porphyromonas gingivalis, an etiological agent in severe forms of periodontitis (a chronic inflammatory disease), is a prominent component of the oral microbiome and a successful colonizer of the oral epithelium. IL-6 and IL-10 were increased with these diseases. These results were a possible use of these cytokines in diagnostic test of periodontitis.
Referencias
Lv, M., Zhou, Y., Polson, S. W., Wan, L. Q., Wang, M., Han, L., ... & Lu, X. L. (2019). Identification of chondrocyte genes and signaling pathways in response to acute joint inflammation.Scientific reports,9(1), 1-12.
Lei, Y., Wang, K., Deng, L., Chen, Y., Nice, E. C., & Huang, C. (2015). Redox regulation of inflammation: old elements, a new story.Medicinal research reviews,35(2), 306-340.
Stankov, S. V. (2012). Definition of inflammation, causes of inflammation and possible anti-inflammatory strategies.Open Inflamm J,5(1), 1-9.
Maruyama, M., Rhee, C., Utsunomiya, T., Zhang, N., Ueno, M., Yao, Z., & Goodman, S. B. (2020). Modulation of the inflammatory response and bone healing.Frontiers in Endocrinology,11, 386.
Frankovich, J., Swedo, S., Murphy, T., Dale, R. C., Agalliu, D., Williams, K., ... & Thienemann, M. (2017). Clinical management of pediatric acute-onset neuropsychiatric syndrome: part IIâ??use of immunomodulatory therapies.Journal of child and adolescent psychopharmacology,27(7), 574-593.
Serhan, C. N., & Savill, J. (2005). Resolution of inflammation: the beginning programs the end.Nature immunology,6(12), 1191-1197.
Jaeschke, H. (2006). Mechanisms of Liver Injury. II. Mechanisms of neutrophil-induced liver cell injury during hepatic ischemia-reperfusion and other acute inflammatory conditions.American Journal of Physiology-Gastrointestinal and Liver Physiology,290(6), G1083-G1088.
Westman, J., Grinstein, S., & Marques, P. E. (2020). Phagocytosis of necrotic debris at sites of injury and inflammation.Frontiers in immunology,10, 3030.
Markiewski, M. M., & Lambris, J. D. (2007). The role of complement in inflammatory diseases from behind the scenes into the spotlight.The American journal of pathology,171(3), 715-727.
Vina, E. R., Fang, A. J., Wallace, D. J., & Weisman, M. H. (2005, December). Chronic inflammatory demyelinating polyneuropathy in patients with systemic lupus erythematosus: prognosis and outcome. InSeminars in arthritis and rheumatism(Vol. 35, No. 3, pp. 175-184). WB Saunders.
Ballini, A., Cantore, S., Farronato, D., Cirulli, N., Inchingolo, F., Papa, F., ... & Scacco, S. (2015). Periodontal disease and bone pathogenesis: The crosstalk between cytokines and porphyromonas gingivalis.J. Biol. Regul. Homeost. Agents,29(2), 273-281.
Skurska, A., Dymicka-Piekarska, V., Milewski, R., & Pietruska, M. (2021). Dynamics of Matrix Metalloproteinase-1 and-8 Secretion in Gingival Crevicular Fluid after Gingival Recession Therapy via MCAT with Either Subepithelial Connective Tissue Graft or Collagen Matrix.Biomolecules,11(5), 731.
Kinane, D. F., Stathopoulou, P. G., & Papapanou, P. N. (2017). Periodontal diseases.Nature Reviews Disease Primers,3(1), 1-14.
Lamont, R. J., Koo, H., & Hajishengallis, G. (2018). The oral microbiota: dynamic communities and host interactions.Nature Reviews Microbiology,16(12), 745-759.
Hajishengallis, G., & Korostoff, J. M. (2017). Revisiting the Page & Schroeder model: the good, the bad and the unknowns in the periodontal host response 40 years later.Periodontology 2000,75(1), 116-151.
Hawiger, J., & Zienkiewicz, J. (2019). Decoding inflammation, its causes, genomic responses, and emerging countermeasures.Scandinavian journal of immunology,90(6), e12812.
Lechner, J., Noumbissi, S., & von Baehr, V. (2018). Titanium implants and silent inflammation in jawboneâ??a critical interplay of dissolved titanium particles and cytokines TNF-a and RANTES/CCL5 on overall health?.EPMA Journal,9(3), 331-343.
Han, Y., Huard, A., Mora, J., da Silva, P., Brüne, B., & Weigert, A. (2020). IL-36 family cytokines in protective versus destructive inflammation.Cellular Signalling, 109773.
Baron, E. J., Peterson, L. R. and Finegoldens, S. M. (1994). Bailey and Scott's diagnostic microbiology. 9th Ed. Mosology. Co. USA.
MacFadden, J. F., (2000). Biochemical tests for identification of medical bacteria. 3rd ed. ed. Baltimore Md: Lippincott Williams and Wilkins.
Marin, M. J., Ambrosio, N., Herrera, D., Sanz, M., & Figuero, E. (2018). Validation of a multiplex qPCR assay for the identification and quantification of Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis: in vitro and subgingival plaque samples.Archives of oral biology,88, 47-53.
Fu, L. H., Cong, B., Zhen, Y. F., Li, S. J., Ma, C. L., Ni, Z. Y., ... & Yao, Y. X. (2007). Methylation status of the IL-10 gene promoter in the peripheral blood mononuclear cells of rheumatoid arthritis patients.Yi Chuan= Hereditas,29(11), 1357-1361.
Riep, B., Edesi-NeuÃ?, L., Claessen, F., Skarabis, H., Ehmke, B., Flemmig, T. F., ... & Moter, A. (2009). Are putative periodontal pathogens reliable diagnostic markers?.Journal of clinical microbiology,47(6), 1705-1711.
Kharitonova, M., Vankov, P., Abdrakhmanov, A., Mamaeva, E., Yakovleva, G., & Ilinskaya, O. (2021). The composition of microbial communities in inflammatory periodontal diseases in young adults Tatars.AIMS microbiology,7(1), 59.
Mysak, J., Podzimek, S., Sommerova, P., Lyuya-Mi, Y., Bartova, J., Janatova, T., ... & Duskova, J. (2014). Porphyromonas gingivalis: major periodontopathic pathogen overview.Journal of immunology research,2014.
Olsen, I., & Singhrao, S. K. (2018). Importance of heterogeneity in Porhyromonas gingivalis lipopolysaccharide lipid A in tissue specific inflammatory signalling.Journal of oral microbiology,10(1), 1440128.
Hasturk, H., & Kantarci, A. (2015). Activation and resolution of periodontal inflammation and its systemic impact.Periodontology 2000,69(1), 255-273.
Gonzales, J. R., Groeger, S., Johansson, A., & Meyle, J. (2014). T helper cells from aggressive periodontitis patients produce higher levels of interleukin-1 beta and interleukin-6 in interaction with Porphyromonas gingivalis.Clinical oral investigations,18(7), 1835-1843.
Atanasova, K. R., & Yilmaz, Ã?. (2014). Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association.Molecular oral microbiology,29(2), 55-66.
Hajishengallis, G., Chavakis, T., & Lambris, J. D. (2020). Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy.Periodontology 2000,84(1), 14-34.
Ceccarelli, F., Saccucci, M., Di Carlo, G., Lucchetti, R., Pilloni, A., Pranno, N., ... & Polimeni, A. (2019). Periodontitis and rheumatoid arthritis: the same inflammatory mediators?.Mediators of inflammation,2019.