Full Length Research Article - (2022) Volume 17, Issue 5
Molecular Similarly Between Infectious Bronchitis Viruses and Common Vaccines
Saja Sameer Abbood* and Balqees Hassan Ali*Correspondence: Saja Sameer Abbood, College of Veterinary Medicine, University of Baghdad, Iraq, Email:
Received: 29-Sep-2022 Published: 13-Oct-2022
Abstract
Infectious bronchitis is an acute extremely infectious respiratory illness caused by the avian gamma-corona virus. Infection with infectious bronchitis virus predisposes the bird to subsequent bacterial infection, worsening the situation. Infection causes severe morbidity and variable mortality in broilers, as well as a significant decrease in layer production of eggs. Samples were collected from clinical cases submitted for necropsy at local veterinary clinics.This study was conducted to detect the molecular similarity in S1 gene sequence between field viruses and commonly used vaccines. In order to compare the sequences of field viruses with vaccinal viruses, two vaccines are chosen based on their popularity in veterinary clinics. These are MA5 strain and H120 strain. Molecular identification was done by using polymerase chain reaction (PCR) which was employed using primers target the S1 gene. Four positive field cases and two vaccine samples were sent to sequencing. The results of sequence alignment showed that vaccine viruses differ by more than 30% when compared to sequences of all the field viruses. The difference between genetic sequence leads to vaccine failure due to difference in the antigenic molecules on the spike protein of IBV.
Keywords
Sport psychology. Sport exercise. Infectious bronchitis virus. Chickens. Sequence identity. vaccines. S1 gene
Introduction
Infectious bronchitis is an acute extremely infectious respiratory illness caused by the avian gamma-corona virus. Chickens and other avian species can be infected with IBV (OIE, 2018). Infection with infectious bronchitis virus predisposes the bird to subsequent bacterial infection, worsening the situation. Infection causes severe morbidity and variable mortality in broilers, as well as a significant decrease in layer production of eggs (Ibrahiem, 2016). The virus may be found all around the world and is spread by respiration or direct bird-tobird contact or exposure to contaminated equipment, litter, tools, or more premises. Although in-ovo spread of the pathogen did not recorded yet, it may contaminate the eggshells by shedding from the reproductive or alimentary system (Jackwood & de Wit, 2020; Mohammed et al., 2013). The virus is an enveloped virus that varies in morphology from round to pleomorphic. The virions are roughly 120 nm in size and have club-shaped outer appendages called spikes and these are approximately 20 nm long, giving the virus a look of a crown.Corona is a latin word means crown (Khataby et al., 2020).
The symptoms of IB in affected young birds include general respiratory signs such as nasal secretion, respiratory rales, coughing, sneezing, and gasping. Watery eyes and even dilated sinuses have been seen in chicks. Other concurrent disease could be contributing to the severity of some cases (Ellakany et al., 2019; Hassan et al., 2017;Khamas, 2008). The chicks could be spotted curled up next to a heat source and seem depressed. Feed intake and growth might both be decreased. If the flock is not properly investigated, the sickness may even go undetected (Jackwood & de Wit, 2020). Even in situations with evident production reductions and the laying of bleached eggs, respiratory abnormalities in laying chickens might be absent or extremely slight. The degree of the production reduction can range from minor to severe, depending on parameters such as the viral serotype and birds immunity, the lay phase during which the infection occurred, and concurrent infections (Najimudeen et al., 2020). The trachea, nasal cavity, and sinuses of affected hens contain exudate. During the acute infection, the air sacs may be frothy, then turbid and have a yellow caseous discharge. Inflammation of lung tissues that surround big bronchi. Infections with strains of renal tropism can cause enlarged, faint kidneys due to the occupation of tubules with urate (Benyeda et al., 2009; Ziegler et al., 2002).
Vaccinated or recently infected poultry are resistant to infection with the same virus strain, while immunity against infection with different IBV strains is variable. The challenge of vaccinated birds with a homologous virus (same strain) leads to much less viral shedding and for a limited duration than in unprotected birds (Vagnozzi et al., 2010). For IBV vaccination, attenuated and killed vaccines are employed. For priming of breeders and layers, live vaccinations are employed and they are also used for broilers. Attenuation is done by repetitive passage in chicken embryos, occasionally in conjunction with thermal processing (Jackwood et al., 2010). There are different vaccine strains used for the imunization of birds against IBV. These include: the Massachusetts strains (Mass41 and H120) (Jackwood & de Wit, 2020), Arkansas (Ark), Connecticut (Conn), Delaware (Del), Georgia98 (GA98), Georgia 08 (GA08), and Georgia 13 (GA13) in the United states, and 793/B, QX, and Q1 in Europe, Asia, and South America (Jordan, 2017). In Iraq, multiple vaccines strains are used including: H 120, MA5, 4/91, QX, Variant2, D274 and M48 (Abdulmaged, 2017; Ali Ameen & Hussein Raoof, 2013; Al-Khafaji, 2013; AL-Zuhariy, 2017; Hammadi & Zahid, 2015; Kadhym & Zahid, 2017; Saood & Al-Mayah, 2017; Zahid et al., 2011).
Our study focused on S1 gene sequence because a few changes in the amino acid sequence could lead to the emergence of a new virus strain (Cavanagh, 2007). When there is new strains, meanwhile, vaccine producers and farm vaccination programs are still relaying on old vaccinal strains will lead to lack of immunological protection of the bird. Hence, this will result in IB outbreaks even in vaccinated birds (Y. Ennaji et al., 2020).
Materials and methods
Sampling
Samples were collected from clinical cases submitted for necropsy at local veterinary clinics. Tissues for molecular detection included tracheas, lungs and kidneys. Those organs were placed on petri dishes and small pieces were cut and put in 1.5 ml microcentrifuge tubes covered with TRIzol™ Reagent and kept in the freezer then sent to PCR laboratories.
Vaccines
In order to compare the sequences of field viruses with vaccinal viruses, two vaccines are chosen based on their popularity in veterinary clinics. These are MA5 strain and H120 strain. Those vaccines were sent for molecular detection along with tissue samples.
Molecular Detection
Polymerase chain reaction (PCR) was employed using primers designed by Raoof et al., (2021). These primers include the forward primer f-IBV-S1 5`- GTT TAC TAC TAC CAA AGT GCC TT -3` and the reverse primer 5`- GTG TAA ACA AGG TCA CCA TTT A -3`. Those oligonucleotides target the S1 gene and produce a 448bp PCR product.
Sequencing and Sequence Analysis
for sequencing step, PCR products were sent to Macrogen Co., Seoul, Republic of Korea. Four positive field cases and two vaccine samples were sent to sequencing. Once the sequence was ready the company emailed the sequence in FASTA format.
To analyse the sequences, two programs were used. The program Geneious Prime was used to generate the sequence similarity percentage and table between the sequences of our study between each other. BLAST® was used to compare the study sequences with sequences of other similar viruses on the NCBI GenBank. The later software was also used to create phylogenetics trees.
Results and Discussion
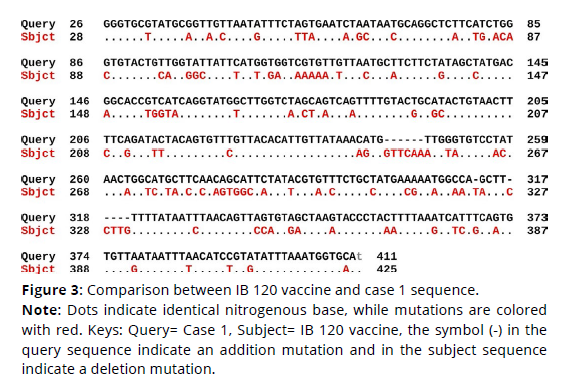
Sequence alignment of IB 120 vaccine virus sequence and the four cases sequences revealed that the percent identity of the IB 120 vaccine sequence and case 1 sequence was 69.85%. The exact differences in the sequences is detailed in Figure 3 (Figures 1-3). There was 108 substitution mutations and 12 addition mutations. On the other hand, case 2 sequence was 69.58% identical to IB 120 sequence. There was 110 substitution mutations and 13 addition mutations (Figure 4). Likewise, The percent identity of the IB 120 vaccine sequence and case 3 sequence was 69.52%. There was 110 substitution mutations, 16 addition mutations and 2 deletion mutations (Figure 5). Moreover, case 4 sequence was 68.77% identical to IB 120 sequence. There was 111 substitution mutations, 12 addition mutations and one deletion mutations (Figure 6).
Figure 2. Comparison between IB 120 and MA5 vaccine sequences.
Note: Dots indicate identical nitrogenous base while mutations are colored
with red. Keys: Query= MA 5 vaccine, Subject= IB 120 vaccine, the symbol (-) in
the query sequence indicate an addition mutation and in the subject sequence
indicate a deletion mutation.
Figure 3. Comparison between IB 120 vaccine and case 1 sequence.
Note: Dots indicate identical nitrogenous base, while mutations are colored
with red. Keys: Query= Case 1, Subject= IB 120 vaccine, the symbol (-) in the
query sequence indicate an addition mutation and in the subject sequence
indicate a deletion mutation.
Figure 4. Comparison between IB 120 vaccine and case 2 sequences.
Note: Dots indicate identical nitrogenous base, while mutations are colored
with red. Keys: Query= Case 2, Subject= IB 120 vaccine, the symbol (-) in the
query sequence indicate an addition mutation and in the subject sequence
indicate a deletion mutation.
Figure 5. Comparison between IB 120 vaccine and case 3 sequences.
Note: Dots indicate identical nitrogenous base, while mutations are colored
with red. Keys: Query= Case 3, Subject= IB 120 vaccine, the symbol (-) in the
query sequence indicate an addition mutation and in the subject sequence
indicate a deletion mutation.
Figure 6. Comparison between IB 120 vaccine and case 4 sequences.
Note: Dots indicate similar nitrogenous base, while mutations are colored with
red. Keys: Query= Case 4, Subject= IB 120 vaccine, the symbol (-) in the query
sequence indicate an addition mutation and in the subject sequence indicate
a deletion mutation.
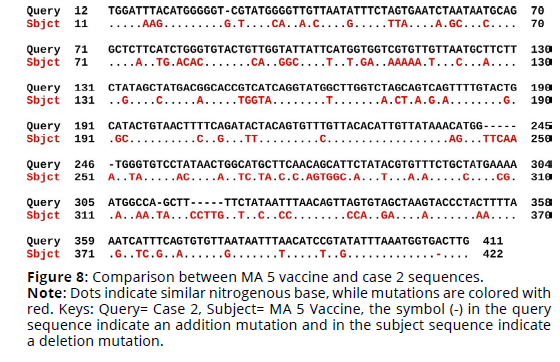
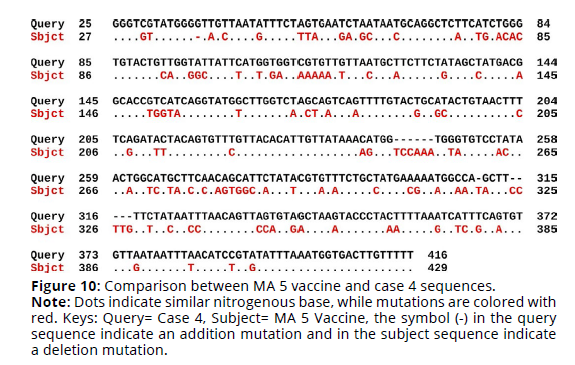
Sequence alignment of MA 5 vaccine virus sequence and the four cases sequences revealed that the percent identity of the MA 5 vaccine sequence and case 1 sequence was 69.42%. The exact differences in the sequences are detailed in Figure 1. There was 110 substitution mutations and 12 addition mutations. On the other hand, case 2 sequence was 69.01% identical to MA 5 vaccine sequence. There was 114 substitution mutations, 13 addition mutations and one deletion mutation (Figures 7, 8). Furthermore, The percent identity of the MA 5 vaccine sequence and case 3 sequence was 68.98%. There was 108 substitution mutations, 15 addition mutations and 2 deletion mutations (Figure 9). Moreover, case 4 sequence was 69.55% identical to MA 5 sequence. There was 110 substitution mutations, 11 addition mutations and one deletion mutations (Figure 10).
Figure 7. Comparison between MA 5 vaccine and case 1 sequences.
Note: Dots indicate similar nitrogenous base, while mutations are colored with
red. Keys: Query= Case 1, Subject= MA 5 Vaccine, the symbol (-) in the query
sequence indicate an addition mutation and in the subject sequence indicate
a deletion mutation.
Figure 8. Comparison between MA 5 vaccine and case 2 sequences.
Note: Dots indicate similar nitrogenous base, while mutations are colored with red. Keys: Query= Case 2, Subject= MA 5 Vaccine, the symbol (-) in the query sequence indicate an addition mutation and in the subject sequence indicate a deletion mutation.
Figure 9. Comparison between MA 5 vaccine and case 3 sequences.
Note: Dots indicate similar nitrogenous base, while mutations are colored
with red. Keys: Query= Case 3, Subject= MA 5 Vaccine, the symbol (-) in the
query sequence indicate an addition mutation and in the subject sequence
indicate a deletion mutation.
Figure 10. Comparison between MA 5 vaccine and case 4 sequences.
Note: Dots indicate similar nitrogenous base, while mutations are colored with
red. Keys: Query= Case 4, Subject= MA 5 Vaccine, the symbol (-) in the query
sequence indicate an addition mutation and in the subject sequence indicate
a deletion mutation.
Sequence alignment of the four cases sequences revealed that case 1 sequence was identical to case 2 sequence by 98.99% identity. The exact differences in the sequences is detailed in Figure 11. There was 4 substitution mutations in case 1 when compared to case 2. In addition, There was 97.84% identity between case 1 and case 3. There was 8 substitution mutations and one addition mutation (Figure 12). Moreover, There was 97.08% identity between case 1 and case 4. There was 9 substitution mutations and 3 deletion mutation (Figure 13). On the other hand, case 2 and case 3 was 99.75% identical with one addition mutation in case 2 sequence when aligned to case 3 sequence (Figure 14). Furthermore, case 2 and case 4 was 98.25% identical. There was 4 substitution mutations, 2 deletion mutations and one addition mutation in case 2 sequence when aligned to case 4 sequence (Figure 15). Finally, case 3 and case 4 was 96.72% identical. There was 10 substitution mutations and 4 deletion mutations in case 3 sequence when aligned to case 4 sequence (Figure 16).
Figure 11. Comparison between case 1 and case 2 sequences.
Note: Dots indicate similar nitrogenous base, while mutations are colored with
red. keys: Query= Case 2, Subject= Case 1, the symbol (-) in the query sequence
indicate an addition mutation and in the subject sequence indicate a deletion
mutation.
Figure 12. Comparison between case 1 and case 3 sequences.
Note: Dots indicate similar nitrogenous base, while mutations are colored
with red. Keys: Query= Case 3, Subject= Case 1, the symbol (-) in the query
sequence indicate an addition mutation and in the subject sequence indicate
a deletion mutation.
Figure 14. Comparison between case 2 and case 3 sequences.
Note: Dots indicate similar nitrogenous base and mutations are colored with
red. Keys: Query= Case 3, Subject= Case 2, the symbol (-) in the query sequence
indicate an addition mutation and in the subject sequence indicate a deletion
mutation.
Figure 15. Comparison between case 2 and Case 4 sequences.
Note: Dots indicate similar nitrogenous base and mutations are colored with
red. Keys: Query= Case 4, Subject= Case 2, the symbol (-) in the query sequence
indicate an addition mutation and in the subject sequence indicate a deletion
mutation.
Figure 16. Comparison between case 3 and case 4 sequences.
Note: Dots indicate similar nitrogenous base and mutations are colored with
red. Keys: Query= Case 4, Subject= Case 3, the symbol (-) in the query sequence
indicate an addition mutation and in the subject sequence indicate a deletion
mutation.
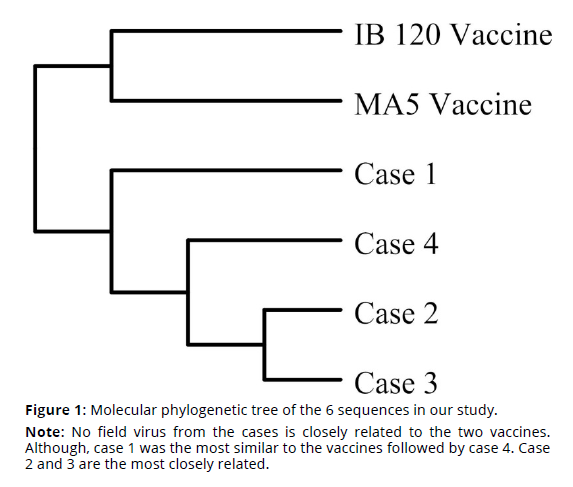
In total, sequence alignment showed that vaccine viruses differ by more than 30% when compared to sequences of all the field viruses (Tables 1-2). Hence, the vaccinal viruses are grouped separately from the rest of field viruses in the molecular phylogenetic tree in Figure 1. This difference is a huge difference in molecular terms especially when taking into consideration that the difference between human and mice genome is only about 20% (Mouse Genome Sequencing Consortium, 2002).
| Virus | GenBank Accession Number |
|---|---|
| IB H120 vaccine | OP373738 |
| MA5 vaccine | OP373739 |
| Case 1 | OP373740 |
| Case 2 | OP373741 |
| Case 3 | OP373742 |
| Case 4 | OP373743 |
| IB 120 Vaccine | MA5 Vaccine | Case 1 | Case 2 | Case 3 | Case 4 | |
|---|---|---|---|---|---|---|
| IB 120 Vaccine | 98.05% | 69.85% | 69.58% | 69.52% | 68.77% | |
| MA5 Vaccine | 98.05% | 69.42% | 69.01% | 68.98% | 69.55% | |
| Case 1 | 69.85% | 69.42% | 98.99% | 97.84% | 97.08% | |
| Case 2 | 69.58% | 69.01% | 98.99% | 99.75% | 98.25% | |
| Case 3 | 69.52% | 68.98% | 97.84% | 99.75% | 96.72% | |
| Case 4 | 68.77% | 69.55% | 97.08% | 98.25% | 96.72% |
Our results emphasize the importance of genetic diversity and confirm the the presence of continuous sequence alterations. This was demonstrated by comparing our sequences to each other and found nucleotide mutations in each pair of isolates compared. The presence of high sequence alterations in IBVs was investigated by Umar et al., (2016). They highlighted the three elements that may cause these alterations. First, There is no RNA polymerase proofreading, this causes errors and hence mutations. Second, the constant use of different live vaccines will lead to recombination and emergence of new strains. Third, continuous circulation of the virus will cause pressure on the birds immune system and increase the chance of errors during replication.
Conclusions
1. There was more than 30% difference in the S1 gene sequence when comparing sequences from vaccines used in Iraq and viruses circulating locally.
2. The difference between genetic sequence leads to vaccine failure due to difference in the antigenic molecules on the spike protein of IBV.
3. Our result showed that gene sequencing provides great benefits in designing and choosing vaccines against local viruses.
4. There was continuous occurrence of mutations in local IBV viruses. This was shown in the comparison between our sequences and previous sequence data in the GenBank.
References
Abdulmaged, S. H. (2017). Efficacy of infectious bronchitis disease vaccines as measured by viral shed after virulent challenge in broiler. Basrah Journal of Veterinary Research., 16(2).
Ali Ameen, N., & Hussein Raoof, H. (2013). Interference of Infectious Bronchitis Virus Vaccine with Immune Response induced by Infectious Bursal Disease Virus Vaccine in Broiler Chicks. Journal of Kerbala University, 9(1), 140–146.
Al-Khafaji, M. A. J. (2013). Evaluation the efficacy of volvac® vaccine to Infectious Bronchitis and determine the best rout of administration. Al-Qadisiyah Journal of Veterinary Medicine Sciences, 12(2), 36–44.
AL-Zuhariy, M. T. (2017). Improved vaccine strategies of infectious bronchitis disease to reduce shedding of virulent virus from infected birds. Kufa Journal For Veterinary Medical Sciences, 8(1).
Benyeda, Z., Mató, T., Süveges, T., Szabo, E., Kardi, V., Abonyi-Tóth, Z., Rusvai, M., & Palya, V. (2009). Comparison of the pathogenicity of QX-like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathology, 38(6), 449–456.
Cavanagh, D. (2007). Coronavirus avian infectious bronchitis virus. Veterinary Research, 38(2), 281–297.
Ellakany, H. F., Elbestawy, A. R., Abd-Elhamid, H. S., Gado, A. R., Nassar, A. A., Abdel-Latif, M. A., Ghanima, I. A., Abd El-Hack, M. E., Swelum, A. A., & Saadeldin, I. M. (2019). Effect of experimental Ornithobacterium rhinotracheale infection along with live infectious bronchitis vaccination in broiler chickens. Poultry Science, 98(1), 105–111.
Ennaji, Y., Khataby, K., & Ennaji, M. M. (2020). Infectious bronchitis virus in poultry: Molecular epidemiology and factors leading to the emergence and reemergence of novel strains of infectious bronchitis virus. In M. M. Ennaji, Emerging and Reemerging Viral Pathogens: Vol. Volume 2: Applied Virology Approaches Related to Human, Animal and Environmental Pathogens (pp. 31–44). Elsevier.
Hammadi, H. A., & Zahid, A. A. H. (2015). Preparation and evaluation of multivalent infectious bronchitis vaccine from commercial vaccine strains. Al-Qadisiyah Journal of Veterinary Medicine Sciences, 14(2), 55–60.
Hassan, K. E., Ali, A., Shany, S. A., & El-Kady, M. F. (2017). Experimental co-infection of infectious bronchitis and low pathogenic avian influenza H9N2 viruses in commercial broiler chickens. Research in Veterinary Science, 115, 356–362.
Ibrahiem, Z. Y. (2016). Molecular detection of Avian Influenza virus, Newcastle disease virus, Infectious bronchitis virus and Mycoplasma gallisepticum in broiler flocks with airsacculitis [M.Sc Thesis]. University of Baghdad.
Jackwood, M. W., & de Wit, S. (2020). Infectious Bronchitis. In D. E. Swayne, M. Boulianne, C. M. Logue, L. R. McDougald, V. Nair, & D. L. Suarez, Diseases of Poultry (pp. 167–188). John Wiley & Sons, Ltd. https://doi.org/10.1002/9781119371199.ch4
Jackwood, M. W., Hilt, D. A., Sellers, H. S., Williams, S. M., & Lasher, H. N. (2010). Rapid heat-treatment attenuation of infectious bronchitis virus. Avian Pathology, 39(3), 227–233.
Jordan, B. (2017). Vaccination against infectious bronchitis virus: A continuous challenge. Veterinary Microbiology, 206, 137–143.
Kadhym, M. J., & Zahid, A. H. (2017) field evaluation of bivalent killed vaccine of infectious bronchitis and newcastle diseases prepared from local isolated strains of broilers in iraq. iraq journal of agricultural research, 22(3).
Khamas, E. J. (2008). Avian influenza (H9N2) outbreak in Iraq. The Iraqi Journal of Veterinary Medicine, 32(1), 223–230.
Khataby, K., Kasmi, Y., Souiri, A., Loutfi, C., & Ennaji, M. M. (2020). Avian Coronavirus: Case of Infectious Bronchitis Virus Pathogenesis, Diagnostic Approaches, and Phylogenetic Relationship Among Emerging Strains in Middle East and North Africa Regions. Emerging and Reemerging Viral Pathogens, 729–744. https://doi.org/10.1016/B978-0-12-819400-3.00033-8
Mohammed, M. H., Hair-Bejo, M., Zahid, bdel A. H., Alazawy, A., Abdul Ahad, E. A., & Hasoon, M. F. (2013). Isolation of Infectious Bronchitis Virus in Primary cells of the Chick Embryo Chorioallantoic Membrane. The Iraqi Journal of Veterinary Medicine, 37(1), 109–114.
Mouse Genome Sequencing Consortium. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature, 420(6915), 520–562. https://doi.org/10.1038/nature01262
Najimudeen, S. M., Hassan, M. S. H., Cork, S. C., & Abdul-Careem, M. F. (2020). Infectious bronchitis coronavirus infection in chickens: Multiple system disease with immune suppression. Pathogens, 9(10), 779.
OIE. (2018). Avian infectious bronchitis. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2018. World Organisation for Animal Health. https://www.oie.int/en/disease/avian-infectious-bronchitis/
Raoof, H. S., Sheikh, M. O. B., & Sulaiman, R. R. (2021). Detection and molecular characterization of infectious bronchitis virus from recent outbreaks in broiler flocks in Sulaimani Governorate. Veterinary Research Forum, 12(1), 117.
Saood, A. I., & Al-Mayah, A. A. (2017). DETECTION OF CILIARY ACTIVITY FOLLOWING VACCINATION WITH 3 COMMERITIAL INFECTIOUS BRONCHITIS VACCINES IN BROILER BIRDS. Infection, 6, 7.
Umar, S., Shah, M. A. A., Munir, M. T., Ahsan, U., & Kaboudi, K. (2016). Infectious bronchitis virus: Evolution and vaccination. World’s Poultry Science Journal, 72(1), 49–60.
Vagnozzi, A., García, M., Riblet, S. M., & Zavala, G. (2010). Protection induced by infectious laryngotracheitis virus vaccines alone and combined with Newcastle disease virus and/or infectious bronchitis virus vaccines. Avian Diseases, 54(4), 1210–1219.
Zahid, A. H., Katam, L. I., & Al-Safy, A. K. (2011). Interfering of infectious bronchitis vaccine (ma5 and 4/91) strains on the immune response of newcastle and avian influenza vaccines in male layers. iraq journal of agricultural research, 2(16).
Ziegler, A. F., Ladman, B. S., Dunn, P. A., Schneider, A., Davison, S., Miller, P. G., Lu, H., Weinstock, D., Salem, M., & Eckroade, R. J. (2002). Nephropathogenic infectious bronchitis in Pennsylvania chickens 1997–2000. Avian Diseases, 46(4), 847–858.